HP Modularity: Difference between revisions
From Santa Fe Institute Events Wiki
| (12 intermediate revisions by 2 users not shown) | |||
| Line 2: | Line 2: | ||
This is the working space for our project. Everything written is a work in progress. | This is the working space for our project. Everything written is a work in progress. | ||
=== Working Draft of Paper === | |||
Below is the current link to the working draft of the paper. I'm not sure if this will really work for version control. Perhaps people can just send me sections and I'll incorporate as needed. -Devin | |||
'''NOT CHECKED OUT''' | |||
[http://www.santafe.edu/events/workshops/images/e/e6/Hp_modularity_v1.doc Link to Word Doc] | |||
[http://www.santafe.edu/events/workshops/index.php/Image:Hp_modularity_v1.doc Link to upload] | |||
=== Title Ideas === | === Title Ideas === | ||
*'''Modularity in coevolving host-pathogen systems''' | |||
*Evolution of modularity in a host-pathogen system | *Evolution of modularity in a host-pathogen system | ||
*Modularity: a pathogen's answer to host interactions | *Modularity: a pathogen's answer to host interactions | ||
| Line 52: | Line 63: | ||
Based on an individual based simulation in matlab that I coded up tonight I got a setup that we can work with while we finalize how we want this thing to work. | Based on an individual based simulation in matlab that I coded up tonight I got a setup that we can work with while we finalize how we want this thing to work. | ||
===== Model Life cycle ===== | |||
#'''Infection''' | #'''Infection''' | ||
##Each host is infected with two different pathogens | ##Each host is infected with two different pathogens | ||
| Line 75: | Line 86: | ||
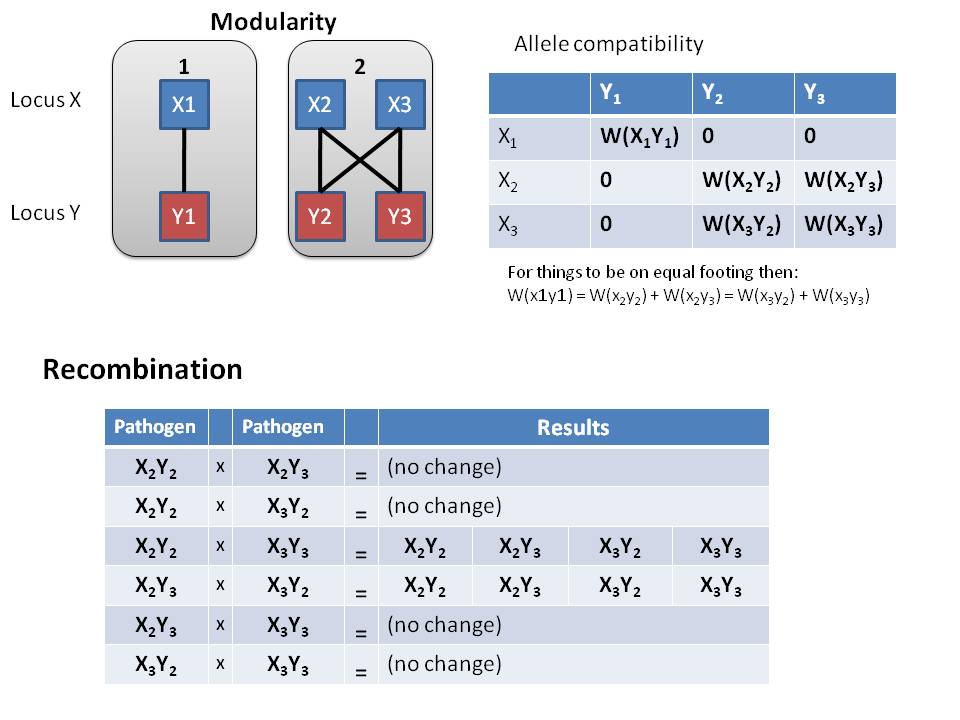
===== Diagram of modularity of alleles in Pathogen ===== | |||
[[Image:Modv4 compat recb.JPG]] | |||
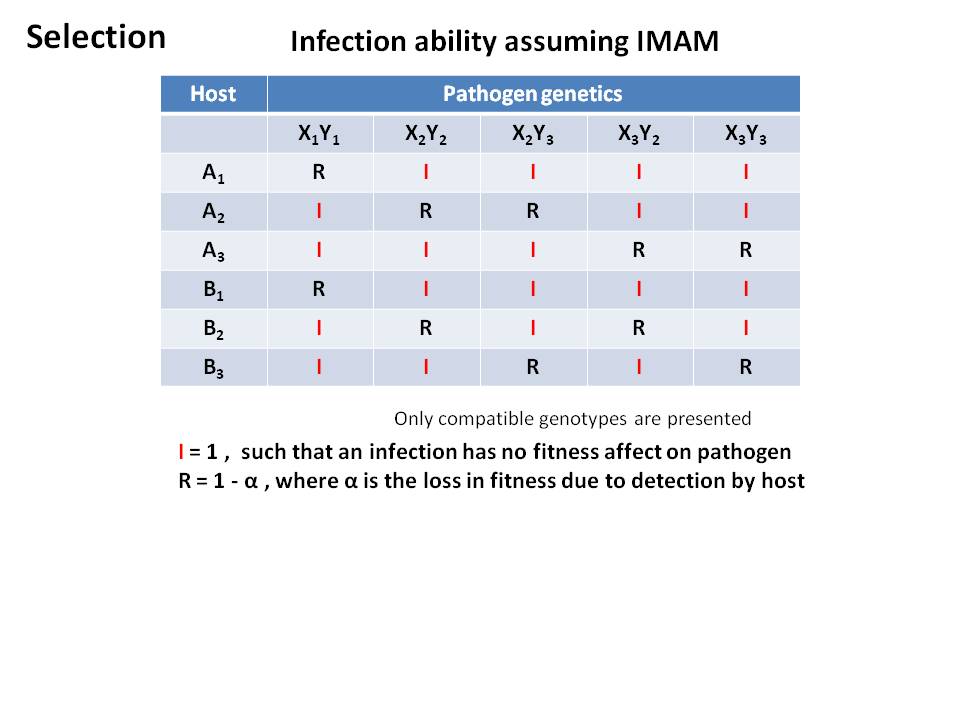
[[Image: | ===== Diagram of interaction between Host and Pathogen ===== | ||
[[Image:Modv4 interation.JPG]] | |||
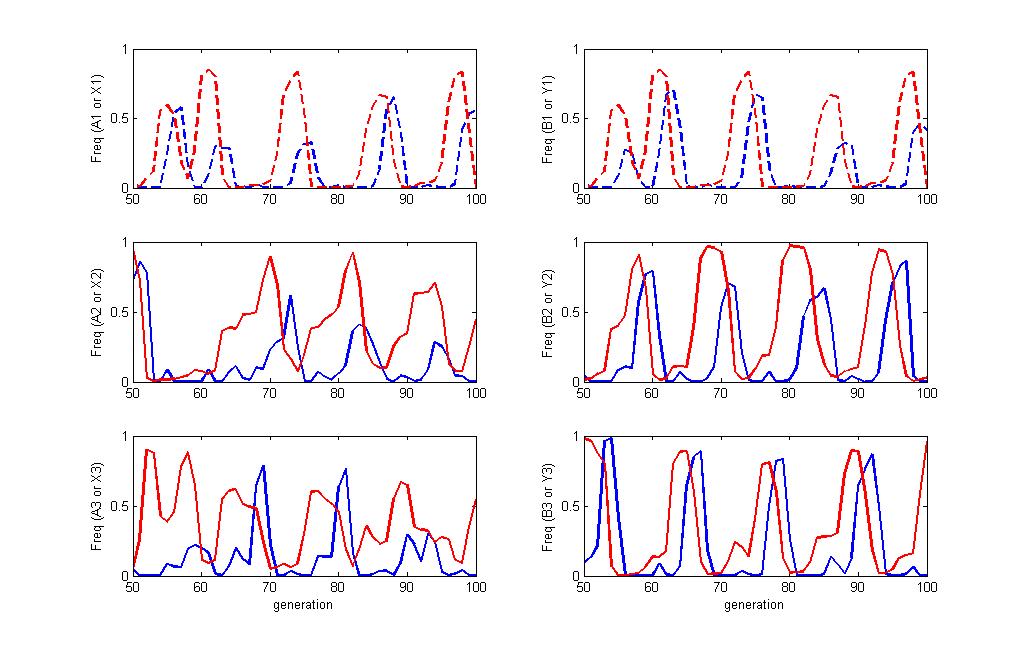
===== Sample dynamics of run ===== | |||
Each panel shows a pair of allele frequencies for the match between the host(blue) and pathogen (red). The top two panels show the "non modular" alleles in dashed lines. All six panels show the same set of time in the simulation but the combinations are pulled out separately for clarity. You can see the frequency dependence of the simulation where the pathogen allele will increase in frequency until a host that can detect it become more frequent at which point, that pathogen will decline. Each suscessive wave of increase cycles through the different alleles in both players. | |||
[[Image:Sampledyn.jpg]] | |||
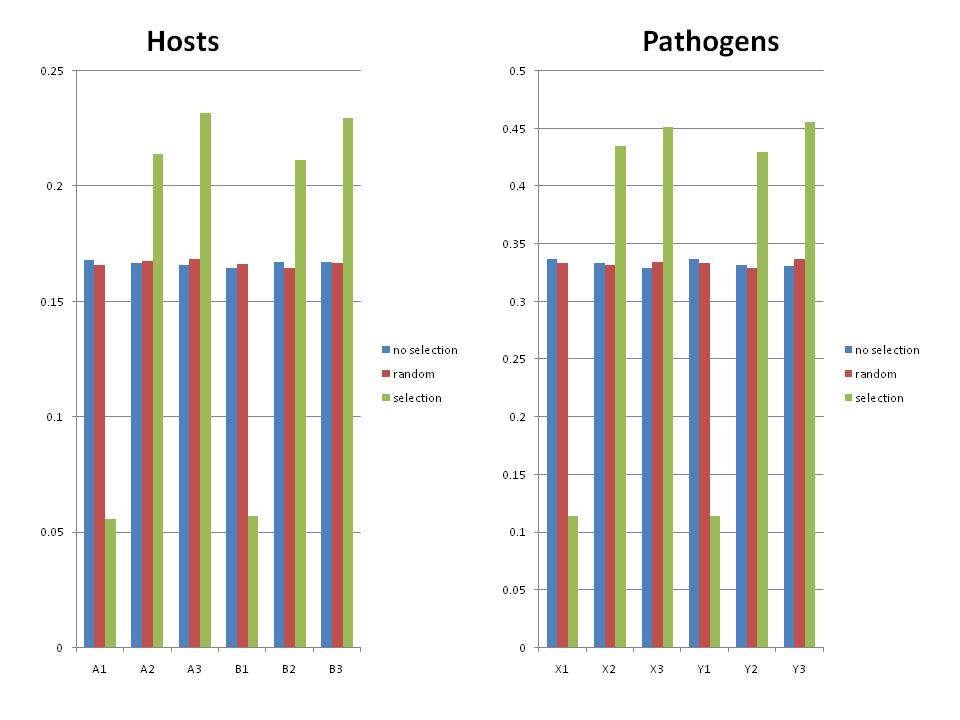
=====Preliminary Results ===== | |||
I ran 3 different scenarios with 1000 host individuals in each case. Each run was started with an equal frequency of each host type at a frequency of 1/6. Each run was started with an equal frequency of each pathogen allele (1/3) and the genotype frequencies were 1/3 for x1y1 and 1/6 for the rest. Run were terminated after a single genotype had fixed or 100 generations (very short time, but most fixed quickly). I ran 10,000 replicates and then collected the mean frenquency of each genotype of host and pathogen at the end of the replicate. Plotted are means and <strike>2x st error of the replicates</strike>. | |||
*'''no selection''' | |||
**Case where there was no fitness interactions between the host and pathogen. Drift alone was working here. | |||
*'''random selection''' | |||
**Case where the host frequencies were randomly assigned each generation, but the frequency of the pathogens was dependent on which infections worked in the previous generation. | |||
**This case simulates a kind of randomly variable environment with no autocorrelation. | |||
**There is no cost to modularity in this case. Because the frequencies hosts are being randomly drawn, then over time, each allele should experience the same selective history and so each should have an equal chance of going to fixation. | |||
**Additional, I found that even if I start with extremely skewed allele frequencies (0.1 of X1, .45 X2, .45 X3), I still get the same final distributions of genotypes ((1/3 of P1 and 1/6 of P2-P5) or equal allele frequencies at the end. In some respects it was more telling to start out with unequal frequencies because we see that the random selection provides correction. | |||
*'''selection''' | |||
**Case where alpha/beta were set to 1, so that an infection was lethal to a host and not infecting was lethal to the pathogen. | |||
**Host and Pathogen frequencies were dependent on the selection each imposed on each other (strong frequency dependence) | |||
[[Image:Modv4 resultsb.jpg]] | |||
==== Replicator equations ==== | |||
This is just one take at trying to get some handle on the replicator dynamics | |||
[http://www.wsu.edu/~drown/modularity_mathematic_v3.html Replicator Dynamics in Mathematica] | |||
== Glossary == | == Glossary == | ||
| Line 130: | Line 170: | ||
Kitano H (2004) Biological robustness. Nature Reviews Genetics 5:826– | Kitano H (2004) Biological robustness. Nature Reviews Genetics 5:826– | ||
837. | 837. | ||
Kreimer A, Borenstein E, Gophna U, Ruppin E (2008) The evolution of modularity in bacterial metabolic networks. Proceedings of the National Academy of Sciences 105:6976-6978. | |||
Lynch M (2007) The frailty of adaptive hypotheses for the origins of organismal complexity. Proceedings of the National Academy of Sciences | Lynch M (2007) The frailty of adaptive hypotheses for the origins of organismal complexity. Proceedings of the National Academy of Sciences | ||
| Line 146: | Line 188: | ||
Welch JJ, Waxman D (2003) Modularity and the Cost of Complexity. Evolution 57:1723-1734 | Welch JJ, Waxman D (2003) Modularity and the Cost of Complexity. Evolution 57:1723-1734 | ||
== Original Discussions == | |||
#'''Impact of host heterogeneity (= consumer-resource dynamics with identity issues)''': ([[Sarah_Cobey|Sarah]] writes:) Most ecological models of consumer-resource interactions assume that all consumers "view" resources the same way, i.e., each resource has only one possible phenotype. For a host-pathogen system, this means that all hosts agree on which strains are identical and which are different, since all hosts are targeting the same antigenic sites (epitopes) of the pathogen in their immune response. When pathogen strains compete in this environment, a broad range of cool dynamics result ([http://www.scienceonline.org/cgi/content/abstract/280/5365/912 Gupta et al., Science, 1998]), depending on the strength of cross-immunity. There is evidence that hosts do not mount identical immune responses when challenged with the same strain of pathogen. In other words, a pathogen's phenotype is a function of the host. How does heterogeneity in hosts' immune responses--this multiplicity of phenotypes--affect competition among pathogens? These could be important results for the field. I'm thinking of doing some simple nonlinear dynamical analysis that builds on the framework in the Gupta paper. This problem seems broadly extensible to antagonistic interactions more generally, but I can't think of specific biological examples. Anyone interested? (Talk to me or post here!) | |||
##Impact of immune system on host-parasite (/pathogen) interaction | |||
##Seems like you could add some details of host genetics and then make up a matrix that describes the fitness dependences of the pathogens for each host genotype. -Devin | |||
##Interesting paper by [http://www.pnas.org/cgi/content/abstract/104/18/7711 Recker et al (2008)] that could be interesting to discuss on this track as well. -Devin | |||
###Can the Recker article be modified by including extra host compartment to represent different host genotypes? | |||
###I'm not sure we need to add extra compartments, especially if we use a status-based approach with the ODEs. It would be worth talking about this problem in front of a blackboard. How about Thursday evening or Sunday? -[[Sarah_Cobey|Sarah]] | |||
####<strike>Let's discuss 6pm Thursday</strike> completed | |||
##Let's get some pictures of what this would this system would look like. I'll try and add something, but other's should drop in a pdf/powerpoint/artististic rendering | |||
#'''Need homes, or just ideas for later''' | |||
##Effects of pathogen competition on epidemic outbreaks | |||
##Spatial heterogeneity of transmission of parasites-- If we have time, I'd love to put the model we have started to develop on a graph or something...see the effects of spatial heterogeneity and the structure of the population on pathogen population, and maybe see how dynamics differ as one moves from an area of homogenous viral population to a "contact zone" where two strains compete...-[[Molly_Rorick|Molly]] | |||
::*<strike>Infectious diseases: epidemic outbreaks vs. endemic steady states</strike> | |||
::*<strike>Direct vs. vector transmission of parasites/pathogens</strike> | |||
::*<strike>Non-genetic transmission of disease resistance</strike> | |||
#First stab at getting some of our ideas in picture form: [[AII_Modularity]] | |||
#This is very interesting. One could imagine using a (weighted?) network picture to capture the transitions. -- [[Jacob_Foster|Jacob]] | |||
###Right now we have a non-weighted network, but expanding the network to allow variation in edge weight will be a nice extension.--[[Molly_Rorick|Molly]] | |||
I'm not at all a biologist, but perhaps there is some way to quantify modularlity in terms of clusters in these networks? Look at cluster dynamics? Clustering that takes into account link weights (e.g. greater link weight than expected by chance)? Perhaps some of this has been looked at? As in http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1865589 -- [[Laura_Feeney|Laura]] | |||
Epidemiology in general. -[[Ruben_Kubiak|Ruben]] | |||
Latest revision as of 04:37, 25 June 2008
Host-Pathogen Modularity
This is the working space for our project. Everything written is a work in progress.
Working Draft of Paper
Below is the current link to the working draft of the paper. I'm not sure if this will really work for version control. Perhaps people can just send me sections and I'll incorporate as needed. -Devin
NOT CHECKED OUT
Title Ideas
- Modularity in coevolving host-pathogen systems
- Evolution of modularity in a host-pathogen system
- Modularity: a pathogen's answer to host interactions
- Do host-pathogen interactions lead to the adaptive evolution for modularity?
- more here
Perspectives on Modularity and Evolvability from the Literature
This section might help to structure our understanding of the general consensus regarding the evolution of modularity. It seems that there are a rather large number of individuals that are skeptical regarding the selective advantages of modularity (Lynch 2007), though it does not appear that any of these individuals have considered the role that host-pathogens may play in determining the benefits associated with modularity, and so we might attempt to motivate our study on this basis.
- "There is no evidence that the modular structure of gene regulatory regions or genetic networks is directly advanced by selective mechanisms. However, the processes of duplication, degenerative mutation, and random genetic drift can lead to the passive emergence of modularity in populations of with genetic effective sizes of the magnitude found in multicellular species" (Lynch 2007, p. 8598).
- Four reasons to question the selective origins of modularity: (i) evolvability is a feature of a population and is thus the focus of group selection, which is less strong than selection acting at the level of the individual; (ii) "[i]t is by no means clear that an enhanced ability to evolve is generally advantageous"; (iii) "there is no evidence that phylogenetic variation in evolutionary features reflects anything more than diversity in variation generating factors that exist for purely physical reasons"; and (iv) "comparative genomics provides no support for the idea that genome architectural changes have been promoted in multicellular lineages so as to enhance their ability to evolve" (Lynch 2007, p. 8603).
- Orr (2000) and Welch and Waxman (2003) have used Fisher's geometric model to study the evolutionary cost of complexity. Both studies concluded, perhaps surprisingly, that this cost is quite high.
Major Questions
- Does modularity in pathogen genes responsible for an interaction with a host increase its ability to adapt to a host?
- under what conditions does pathogen modularity arise? specifically,
- Is modularity in the host required?
- what configurations of the interaction matrix?
- what influence does population structure have (in the hosts)?
- additional ones here
Questions to answer in proposal
- why are we studying host-pathogen interactions?
- what evidence suggests that modular allelic dependencies might be advantageous in such a host-pathogen system?
- give a one-line description of this type of modularity
- what conditions do we aim to test to show a benefit to modular dependencies/interaction structures?
- what are our null models, i.e. why is (this) result a surprise?
- what impact does this have for the understanding of host-pathogen interactions, and evolvability in general?
Proposal Introduction
While interactions between species such as hosts and their pathogens (or plants and pollinators, herbivores and plants) have long been implicated as a means of generating patterns of diversification (Ehrlich and Raven 1964; Thompson 1994, 2005), the process by which forces generate robustness and evolvability is not well understood for coevolutionary systems. Previous authors (citation) have suggested that environmental variation leading to temporal changes in fitness can select for modularity in genetic architecture. In the simplest form, Host-pathogen interactions can lead to autocorrelated variation in fitnesses. However, it remains unclear if this type of variation will generate the same kind of selective forces for modularity. These interactions provide an opportunity to study a different form of temporal variation in fitnesses and allow us to test this potentially strong form of selection.
We propose to test whether interactions between coevolving hosts and pathogens provides the conditions necessary for selection for modularity of the pathogen alleles responsible for the interaction with the host. Here we model an abstract interaction between a host and pathogen. In this paper we will compare the selective advantage of pathogens with modularity against pathogens without modularity. The working hypothesis is that antagonistic coevolutionary interactions generate substantial amounts of autocorrelated temporal variation in selection which selects modularity of the pathogen alleles responsible for the interaction with the host.
Model design: We have modeled an antagonistic interaction with two players (host and pathogen). In this system all individuals are haploid. Hosts have only one locus which is the interaction locus with multiple alleles. Pathogens have two loci. Each locus has three alleles. The fitness outcome of an interaction between a host and pathogen is modified by allele combinations at the interaction loci. We have modeled a straightforward case using a inverse matching allele model for the interaction. A match decreases the pathogen’s fitness and increases the host’s fitness. We take an abstract approach and cast this model in terms of replicator dynamics of each player in the interaction.
Model Details
Discrete or continuous?
- Pros for discrete: easier to run iterated simulations; possible to run experiments on explicit graph structures
- Pros for continuous: mathematical derivations cleaner
Proof of concept
Based on an individual based simulation in matlab that I coded up tonight I got a setup that we can work with while we finalize how we want this thing to work.
Model Life cycle
- Infection
- Each host is infected with two different pathogens
- This allows limited recombination
- Recombination
- Given our compatibility matrix of alleles, there is only 2 combinations that will result in more diversity in the host
- X2Y2 and X3Y3 yields all 4 modular genotypes
- X2Y3 and X3Y2 yields all 4 modular genotypes
- None of the other combination result in increased diversity of pathogens within a host
- Given our compatibility matrix of alleles, there is only 2 combinations that will result in more diversity in the host
- Host-Pathogen fitness interaction
- Given our interaction matrix, a match of any host allele will result in detection by the host
- decrease in fitness of the pathogen
- lack of detection results in decrease in fitness of the host
- Given our interaction matrix, a match of any host allele will result in detection by the host
- Competition of pathogens
- Each of the pathogen genotypes present in the host compete against one another according their fitness after interaction with the host.
- If two pathogens have different fitness, then the one with the greater fitness will dominate that host and exclude the other pathogen.
- If two pathogens have equal fitness, then they will both dominate the host
- Each of the pathogen genotypes present in the host compete against one another according their fitness after interaction with the host.
- Replication
- Hosts contribute the next generation in proportion to their fitness (it can be decreased by a infection)
- Pathogens contribute to the next generation in proportion to their fitness and the number of other genotypes present in a host.
- If there are two genotypes present in a host, then they each contribute half the amount that a single infection would generate given equal fitness.
Diagram of modularity of alleles in Pathogen
Diagram of interaction between Host and Pathogen
Sample dynamics of run
Each panel shows a pair of allele frequencies for the match between the host(blue) and pathogen (red). The top two panels show the "non modular" alleles in dashed lines. All six panels show the same set of time in the simulation but the combinations are pulled out separately for clarity. You can see the frequency dependence of the simulation where the pathogen allele will increase in frequency until a host that can detect it become more frequent at which point, that pathogen will decline. Each suscessive wave of increase cycles through the different alleles in both players.
Preliminary Results
I ran 3 different scenarios with 1000 host individuals in each case. Each run was started with an equal frequency of each host type at a frequency of 1/6. Each run was started with an equal frequency of each pathogen allele (1/3) and the genotype frequencies were 1/3 for x1y1 and 1/6 for the rest. Run were terminated after a single genotype had fixed or 100 generations (very short time, but most fixed quickly). I ran 10,000 replicates and then collected the mean frenquency of each genotype of host and pathogen at the end of the replicate. Plotted are means and 2x st error of the replicates.
- no selection
- Case where there was no fitness interactions between the host and pathogen. Drift alone was working here.
- random selection
- Case where the host frequencies were randomly assigned each generation, but the frequency of the pathogens was dependent on which infections worked in the previous generation.
- This case simulates a kind of randomly variable environment with no autocorrelation.
- There is no cost to modularity in this case. Because the frequencies hosts are being randomly drawn, then over time, each allele should experience the same selective history and so each should have an equal chance of going to fixation.
- Additional, I found that even if I start with extremely skewed allele frequencies (0.1 of X1, .45 X2, .45 X3), I still get the same final distributions of genotypes ((1/3 of P1 and 1/6 of P2-P5) or equal allele frequencies at the end. In some respects it was more telling to start out with unequal frequencies because we see that the random selection provides correction.
- selection
- Case where alpha/beta were set to 1, so that an infection was lethal to a host and not infecting was lethal to the pathogen.
- Host and Pathogen frequencies were dependent on the selection each imposed on each other (strong frequency dependence)
Replicator equations
This is just one take at trying to get some handle on the replicator dynamics
Replicator Dynamics in Mathematica
Glossary
Evolvability Concepts
Some definitions of 'evolvability' drawn from the literature:
- "The capacity of a lineage to evolve has been termed its evolvability, also called evolutionary adaptability. By evolvability, we mean the capacity to generate heritable, selectable phenotypic variation. This capacity may have two components: (i) to reduce the potential lethality of mutations and (ii) to reduce the number of mutations needed to produce phenotypically novel traits" (Kirschner and Gerhart 1998, p. 8420).
Modularity Concepts
I'm quite sure that there is some overloading here; but these are the types of modularity that have come up in our discussions:
- Modularity for reassortment (Molly)
- Modularity for independence (Molly) (I call this 'functional decomposition' - Rob)
- Repeated modularity: eg where a body segment occurs many times in an organism (Rob)
- Modularity for robustness to antagonistic pleiotrophy (Molly)
Some definitions of 'modularity' drawn from the literature:
- "A module...is a part of an organism that is integrated with respect to a certain kind of process...and relatively autonomous with respect to other parts of the organism" (Wagner et al. 2007, p. 921)
- "All ides of modularity refer to a pattern of connectedness in which elements are grouped into highly connected subsets - that is. modules - which are more loosely connected to other such groups" (Wagner et al. 2007, p. 921)
Robustness Concepts
Some definitions of 'robustness' drawn from the literature:
- "[R]obustness is the maintenance of specific functionalities of the system against perturbations, and it often requires the system to change its mode of operation in a flexible way. In other words, robustness allows changes in the structure and components of the system owing to perturbations, but specific functions are maintained" (Kitano 2004, p. 827).
- Epitope : the phenotype of a pathogen that interacts with a host
- only need to consider a portion of the host's genome since the pathogen only targets a specialised portion of that genome.
References
Chen Y, Dokholyan N (2006) The coordinated evolution of yeast proteins is constrained by functional modularity. Trends in Genetics 22:416–419.
D’Auria G, Jim´enez N, Peris-Bondia F, Pelaz C, Latorre A, et al. (2008) Virulence factor rtx in Legionella pneumophila, evidence suggesting it is a modular multifunctional protein. BMC Genomics 9:14.[pdf]
Early DJ, Deem MW (2004) Evolvability is a selectable trait. Proceedings of the National Academy of Sciences 101:11531-11536 [pdf]
Hintze A, Adami C (2008) Evolution of complex modular biological networks. PLoS Computational Biology 4:e23.[pdf]
Kashtan N, Alon U (2005) Spontaneous evolution of modularity and network motifs. Proceedings of the National Academy of Sciences 102:13773– 13778.[pdf]
Kirschner M, Gerhart J (1998) Evolvability. Proceedings of the National Academcy of Sciences 95:8420–8427.[pdf]
Kitano H (2004) Biological robustness. Nature Reviews Genetics 5:826– 837.
Kreimer A, Borenstein E, Gophna U, Ruppin E (2008) The evolution of modularity in bacterial metabolic networks. Proceedings of the National Academy of Sciences 105:6976-6978.
Lynch M (2007) The frailty of adaptive hypotheses for the origins of organismal complexity. Proceedings of the National Academy of Sciences 104:8597–8604.[pdf]
Orr, HA (2000) Adaptation and the cost of complexity. Evolution 54:13-20 [pdf]
Rives A, Galitski T (2003) Modular organization of cellular networks. Pro- ceedings of the National Academy of Sciences 100:1128–1133.[pdf]
Schlosser G, Wagner G (2004) Modularity in Development and Evolution. Chicago: University of Chicago Press.
Wagner G, Pavlicev M, Cheverud J (2007) The road to modularity. Nature Reviews Genetics 8:921–931.
Welch JJ, Waxman D (2003) Modularity and the Cost of Complexity. Evolution 57:1723-1734
Original Discussions
- Impact of host heterogeneity (= consumer-resource dynamics with identity issues): (Sarah writes:) Most ecological models of consumer-resource interactions assume that all consumers "view" resources the same way, i.e., each resource has only one possible phenotype. For a host-pathogen system, this means that all hosts agree on which strains are identical and which are different, since all hosts are targeting the same antigenic sites (epitopes) of the pathogen in their immune response. When pathogen strains compete in this environment, a broad range of cool dynamics result (Gupta et al., Science, 1998), depending on the strength of cross-immunity. There is evidence that hosts do not mount identical immune responses when challenged with the same strain of pathogen. In other words, a pathogen's phenotype is a function of the host. How does heterogeneity in hosts' immune responses--this multiplicity of phenotypes--affect competition among pathogens? These could be important results for the field. I'm thinking of doing some simple nonlinear dynamical analysis that builds on the framework in the Gupta paper. This problem seems broadly extensible to antagonistic interactions more generally, but I can't think of specific biological examples. Anyone interested? (Talk to me or post here!)
- Impact of immune system on host-parasite (/pathogen) interaction
- Seems like you could add some details of host genetics and then make up a matrix that describes the fitness dependences of the pathogens for each host genotype. -Devin
- Interesting paper by Recker et al (2008) that could be interesting to discuss on this track as well. -Devin
- Can the Recker article be modified by including extra host compartment to represent different host genotypes?
- I'm not sure we need to add extra compartments, especially if we use a status-based approach with the ODEs. It would be worth talking about this problem in front of a blackboard. How about Thursday evening or Sunday? -Sarah
Let's discuss 6pm Thursdaycompleted
- Let's get some pictures of what this would this system would look like. I'll try and add something, but other's should drop in a pdf/powerpoint/artististic rendering
- Need homes, or just ideas for later
- Effects of pathogen competition on epidemic outbreaks
- Spatial heterogeneity of transmission of parasites-- If we have time, I'd love to put the model we have started to develop on a graph or something...see the effects of spatial heterogeneity and the structure of the population on pathogen population, and maybe see how dynamics differ as one moves from an area of homogenous viral population to a "contact zone" where two strains compete...-Molly
Infectious diseases: epidemic outbreaks vs. endemic steady statesDirect vs. vector transmission of parasites/pathogensNon-genetic transmission of disease resistance
- First stab at getting some of our ideas in picture form: AII_Modularity
- This is very interesting. One could imagine using a (weighted?) network picture to capture the transitions. -- Jacob
- Right now we have a non-weighted network, but expanding the network to allow variation in edge weight will be a nice extension.--Molly
I'm not at all a biologist, but perhaps there is some way to quantify modularlity in terms of clusters in these networks? Look at cluster dynamics? Clustering that takes into account link weights (e.g. greater link weight than expected by chance)? Perhaps some of this has been looked at? As in http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1865589 -- Laura
Epidemiology in general. -Ruben