Information Theory of the Heart Page
From Santa Fe Institute Events Wiki
Contact: Hiroshi (hashika1@jhmi.edu)
Thank you for your interest in the project. Feel free to join us any time and share your thoughts!
06/12/2014(Thu) Brainstorming Session
Why: To identify adaptive mechanisms leading to cardiac arrhythmia in diseased hearts. Then localize the origin of arrhythmia (= wavebreak) and treat it before it happens.
Hypothesis: Arrhythmias result from an adaptive mechanism to optimize information transmission in abnormal hearts.
Challenge: Wavebreaks can also occur in normal heart tissue. Would information theory metrics be sensitive enough?
How: Time series from a cellular automata model of the 2-D heart tissue. ?Time series of invasive electrogram from animal or human
What: To quantify information transmission within the heart
Potential Metrics
Mutual information of binary time series (0–resting, 1–excited) in two locations. A high mutual information suggests electrical coupling.
Permutation Entropy - Temporal uncertainty in one location. Time series in rational numbers.
Transfer Entropy - Spatial uncertainty between two locations. Time series in binary numbers.
06/13/2014(Fri) Unofficial Brainstorming Session
Thank you all for suggesting excellent reference articles.
1. Cao et al., Detecting Dynamical Changes in Time Series Using the Permutation Entropy. File:2004 CaoY PRE.pdf - Application of Permutation Entropy to EEG data
2. Rosso et al., Distinguishing Noise from Chaos. File:2007 RossoOA PRL.pdf - Complexity–Entropy Causality plane
3. Lacasa et al., From Time Series to Complex Networks: The Visibility Graph. File:2008 LacasaL PNAS.pdf
4. Lizier et al., Local Information Transfer as a Spatiotemporal Filter for Complex Systems. File:2008 LizierJT PRL.pdf - Local Transfer Entropy
5. Cheong et al., Information Transduction Capacity of Noisy Biochemical Signaling Networks. File:2011 Cheong Science.pdf - Not quite relevance but important for future applications.
Let's read the reference papers above this weekend and discuss what exactly we should measure at the next meeting. The next meeting will be at 02:30pm on Monday 06/16/2014 (after my tutorial[1]) in the lecture hall. If you have any questions I will be around this weekend. You will most certainly find me in the common room (or wherever TV is available) at 7pm on Saturday 06/14/2014 for FIFA World Cup Japan vs. Côte d'Ivoire!
06/15/2014(Sun) Group Meeting
Meeting with Josh @11:00am to discuss the project. Location: ARIEL
 .
.
Thank you everyone for great discussions!
Summary of Information Theory Indices to Quantify Information Transfer within the Heart
Question: So which one would help identify the location of wavebreaks (= intersection between wavefront and wavetail)?
1. Mutual Information 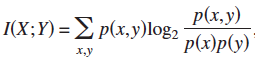 ;
; ![]()
2. Conditional Mutual Information ![]()
3. Transfer Entropy (from state Y to X) 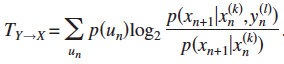 ;
; ![]()
= Conditional Mutual Information regarding the next state X' ![]()
= Global average of a Local Transfer Entropy at each observation ![]()
4. Local Transfer Entropy 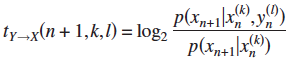 ; Binary time series.
; Binary time series.
See Lizier File:2008 LizierJT PRL.pdf and his recent book chapter (in dropbox)[2]
5. Permutation Entropy ![]() where the sum runs over all n! permutations pi of order n. Real-value time series.
where the sum runs over all n! permutations pi of order n. Real-value time series.
See Bandt and Pompe File:2002 Bandt PRE.pdf and Cao File:2004 CaoY PRE.pdf
6. Complexity-Entropy Causality Plane. See Rosso File:2007 RossoOA PRL.pdf
7. Visibility Graph: A simple rule to map real-valued time series into a network. See Lacasa File:2008 LacasaL PNAS.pdf
8. Graph Entropy: Quantifies structural information of a graph based on a derived probability distribution. See Dehmer File:2008 Dehmer AMC.pdf
9. Brian's time series analysis method: ref?
Project Milestones
I arbitrarily created 3 milestones to split the group into 3 subgroups so everyone can contribute to one aspect of the project. However, at this stage you could consider these milestones a rough idea; I am sure there will be more to do!
1. Build a 2-D cellular automata (CA) model of realistic heart tissue
Available Resources:
- Published model File:2005 Atizenza.pdf
- Netlogo CA code (in dropbox); Original code is here[3]
- Matlab Central: Spiral waves (non-CA)[4]
2. Extract time series from the CA model, modify the existing codes to calculate information theory indices specific to the CA model.
Available Resources:
- Netlogo CA code (in dropbox); Original code is here[5]
- Transfer Entropy Toolbox: Matlab code [6]; ref File:2011 ItoS PLOS.pdf
- Information Dynamics Toolkit: Java code (usable in Matlab) for local transfer entropy, etc. [7]
- Permutation Entropy: Matlab code [8]; ref File:2013 RiedlM EPJST.pdf
- Visibility Graph: Matlab code [9]
3. Statistical analysis, data visualization and communication
Available Resources:
- Lizier's book chapter (in dropbox)[10]
- To identify the locations of wavebreaks, what's the length of the time series data we need to obtain per observation? How can we justify it?
- How many times do we need to repeat observations (the model is not entirely deterministic) to make the data statistically robust?
- What kind of statistical metrics do we use for the final data analysis?
- What kind of results are anticipated?
- How can we visualize the data to effectively communicate the results to the non-expert audience?
The next meeting will be at 02:30pm on Monday 06/16/2014 (after my tutorial[11]) in the lecture hall. Let's think about which milestone you want to contribute to and discuss it.
06/16/2014(Mon) Group Meeting @02:30pm in the Lecture Hall
1. Model Coding - Degang, José, Shai, Hiroshi
Progress:
- Went over the algorithm of Atienza CA model File:2005 Atizenza.pdf
- Emailed the corresponding author to request the source code (Matlab)
- Will start coding the model in Python tomorrow
- Or if we get lucky, we may receive the source code for the model!
2. Data Analysis - Brian, Beth, Shai, Flavia
Progress:
- Extracted cell-by-cell binary time series from the simple Netlogo model
- Discussed the workflow of the data analysis
Unanswered Questions:
- Binary time series data - Should it represent ONLY deporalization (beginning of excitation = 1, otherwise zero), or the entire action potential duration (APD) (from the beginning of excitation to the end of recovery = 1, otherwise zero)? To identify wavebreaks it may be necessary to incorporate the entire APD?
3. Statistical Analysis and Data Visualization - Bernardo, Degang, José, Shai, Brian, Beth, Flavia, Hiroshi
Progress:
- Pending data analysis
- Data Visualization : Local information dynamics indices at each time step at each spatial location -> Aggregate indices at each spatial location?
- Build a directed, weighted network structure based on pairwise local information dynamics indices.
The next meeting will be at 03:00pm on Tuesday 06/17/2014 in the lecture hall. See you then!
06/17/2014(Tue) AM Group Meeting 10:30am-12:00pm in Ariel
1. Model Coding - Degang, José, Shai, Hiroshi
- Went over the math behind a reaction-diffusion model of the 2-D cardiac tissue (Dr. Peter Hammer, Havard Biorobotics Lab) [12]
- Obtained permission from Dr. Hammer to use his model (Thanks, Dr. Hammer!)
- References:
1. File:1993 RogersJM IEEE.pdf
2. File:1993 Pertsov CircRes.pdf
3. FitzHugh-Nagumo model [13]
06/17/2014(Tue) PM Group Meeting 03:00pm-04:30pm in the Lecture Hall
1. Model Coding - Degang, José, Beth, Flavia, Hiroshi
- Continued to go over the math behind Dr. Hammer's model
- Neumann boundary conditions
- Worked on coding to get the time series data from the model
Unanswered Questions:
- The model utilizes the finite difference method to solve PDE - How to optimize it? Time steps? Parameters? Any other tools we should look into? (Matlab PDE toolbox, finite element method, etc.)
- The model outputs real-valued data - what would be an appropriate threshold to binarize real-valued data?
- Hammer's program has been modified and now it (SpiralWaves20140617.m on Dropbox) is able to output binary time series in Brian's sparse format. Modify variable 'threshold' to change the binarization threshold. Modify variable 'dur' to change the length of the time series. Modify variable 'plot_flag' to indicate whether you want the results plotted while running. If you only need the time series, set it to 'false'.
We will have two meetings (again) tomorrow (Wednesday 06/18/2014):
09:00am - 10:30am in the lecture hall
02:00pm - 05:00pm in the lecture hall
See you then!
06/18/2014(Wed) Group Meetings 09:00am-10:30am and 02:00pm-05:00pm in the Lecture Hall
Cellular Automata Model and Data - Brian
- The CA NetLogo model has been greatly improved: removing unnecessary variables and code, simplifying the model, and adding a user interface for simulating different stimulation patterns and printing data to a file. It is on Dropbox as "code/SimHeartCA.nlogo".
- Data has been extracted from the model under various parameter settings, corresponding to (a) a diagonal wave, (b) a radial wave, (c) stable cross-field stimulation, (d) cross-field stimulation with some deviations eventually degenerating into chaotic behavior, and (e) cross-field stimulation immediately degenerating into chaotic behavior. For each dataset, we have extracted the time series for each cell, computed a directed measure of "flow" from node u to v by looking at the average lag time between times when u is excited and times when v is excited, and created a visualization in GraphML format as well as matrices of x- and y-components of the flow vectors. All of this is on Dropbox in the "data" folder.

